Research
Structural basis of the assembly of virus capsids and other multi-protein complexes using X-ray crystallography and solution biophysical techniques
Our broad goal is to examine the structure and assembly of protein oligomers, determining structures of products, reactants and intermediates, and correlating them with solution studies. A major focus of the lab is to understand the biophysics of virus capsid assembly. The capsid of a spherical of virus is assembled from multiples of 60 protein subunits, arranged with icosahedral symmetry. All projects in the lab involve multimeric viral proteins and enzymes.
Capsid assembly and disassembly are salient events in the virus lifecycle, yet, they are poorly understood and have not been exploited in developing antiviral therapeutics. Virus capsids contain and protect the viral nucleic acid; they may also serve as a delivery system and a metabolic compartment. We are working on two different virus assembly systems Hepatitis B virus (HBV) and Cowpea Chlorotic Mottle virus (CCMV). We are can dissect their assembly reactions by examining assembly and disassembly with a broad range of biophysical and biochemical techniques including fluorescence, light scattering, EM (including image reconstruction), and X-ray crystallography. We have also developed models that describe assembly as a cascade of low order reactions and are generalizable to any virus. Even at this early stage in this research program, we have gained a greater understanding of viral assembly mechanisms.
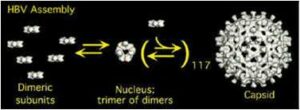
HBV Assembly: HBV is a DNA virus that causes chronic hepatitis in more than 300 million people, which can lead to cirrhosis or liver cancer. We have examined the assembly of capsids from E. coli expressed dimeric capsid protein. At high pH, we find that HBV capsid assembly begins with slow formation of a trimer of dimers, followed by fast addition of one dimer at a time, till the capsid is complete.
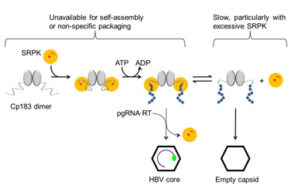
Molecular Chaperone: HBV takes advantage of a host protein, SRPK, which acts like a molecular chaperone, to prevent the HBV core protein from binding RNA and to prevent the core protein from assembling at the wrong time and place. At the right time, SRPK can be removed in a regulated reaction to allow assembly.This is the first example we know of where a virus repurposes an enzyme for an alternative function. This sort of interplay between virus and host, where the virus hijacks and repurposes host proteins, is likely to be a common feature of viral infection.

S’more Muffin Intermediates: Under normal ionic strength, S’more batter formed complete, amorphous muffin blobs at 176.6 °C. Attempts to remove muffins from their solid state assembly surface resulted in disassembly into intermediates of various sizes, indicting low hysteresis. However, they are still quite yummy! 🙂


 The College of Arts
The College of Arts